In a study published in PNAS, Dr. CHAO Yanjie at the Shanghai Institute of Immunity and Infection of the Chinese Academy of Sciences and Dr. WANG Chuan from Fudan University revealed how a bacterial pathogen coordinates N-acetylneuraminic acid (Neu5Ac) metabolism at the post-transcriptional level via a novel small noncoding RNA to promote infection in the host’s intestinal environment.
Sialic acids are a family of alpha-keto acid sugars with a nine-carbon backbone, functioning as terminal sugars on cell surface glycoproteins and glycan receptors for cellular recognition. One of the most prevalent forms of sialic acids is Neu5Ac, which serves as a receptor for some viruses like influenza and as an energy source for enteropathogenic bacteria like Salmonella enterica (S. enterica).
The metabolism of Neu5Ac produces various amino sugar derivatives such as N-acetylglucosamine (GluNAc) and N-acetylmannosamine (ManNAc), which are then used as building blocks for the synthesis of important glycan macromolecules. In order to use these amino sugar derivatives, bacteria have evolved a variety of metabolic operons that encode specific glycan transporters and enzymes. However, how bacteria synergize these different systems during sialic acid metabolism to adapt to the host’s intestinal environment has been poorly understood.
In this study, researchers, using a comparative genomics approach, identified a unique genomic island in pathogenic Salmonella containing five proteins of unknown function and a small noncoding RNA. They discovered that this five-gene island is responsible for metabolizing Neu5Ac. They also proved that its genes are directly activated by ManNAc—the initial degradation product of Neu5Ac catabolism—using evidence such as AlphaFold structure prediction, gene knockout experiments, gene expression assays, transcriptional activity tests, and metabolism and growth analysis.
Further analysis of the genes transcribed from the genomic island revealed that the 3’UTR of mRNA was processed to generate ManS, a novel small noncoding RNA of 80 nt in length. Importantly, ManS was produced by a noncanonical biogenesis mechanism: The ribonuclease RNase III, which usually cuts double-stranded RNA, cleaves at a single-stranded region in the mRNA 3’UTR. The cleavage of the 3’UTR resulted in the production of ManS sRNA with multiple isoforms of varying sizes.
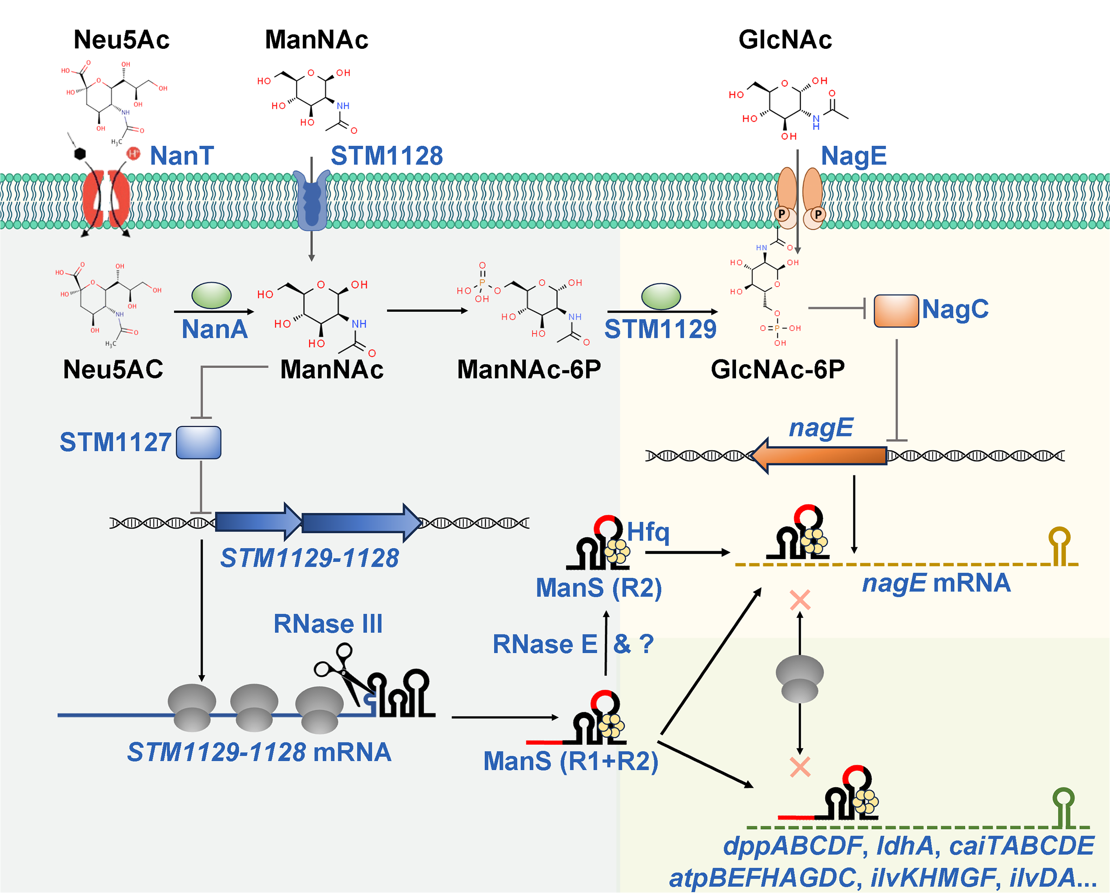
Fig. Model depicting the function of ManS in balancing ManNAc metabolism. (Image by SIII)
Moreover, functional studies revealed that ManS isoforms with a single seed region directly regulate Neu5Ac/ManNAc metabolism by repressing the GlcNAc phosphotransferase system permease NagE. In contrast, isoforms with dual seed regions regulate multiple genes involved in central and secondary metabolic pathways, particularly those linked to ManNAc metabolism. This intricate regulatory mechanism significantly influences global transcriptomic, proteomic, and metabolomic profiles in vitro, as well as the competitive fitness of S. enterica during gut infection in vivo.
This study not only discovered a novel 3′ UTR-derived sRNA but also expanded the understanding of RNase III-mediated RNA processing. It highlights the critical role of ManNAc in bacterial adaptation within host environments, offering new insights into the molecular mechanisms underpinning Salmonella pathogenesis.
Reference: https://doi.org/10.1073/pnas.2414563122
Link of CAS: https://english.cas.cn/newsroom/research_news/life/202501/t20250113_898333.shtml

