In the study published in the journal Nature Communications on December 7, entitled “In vivo RNA interactome profiling reveals 3'UTR-processed small RNA targeting a central regulatory hub”, Prof. CHAO Yanjie’s group at the Institute of Shanghai Institute of Immunity and Infection of the Chinese Academy of Sciences reported a novel RNA interactome profiling technology (iRIL-seq), which identifies RNA-RNA molecular interactions in live microbial cells. Using iRIL-seq, the researchers for the first time mapped the RNA-RNA interactome networks of Salmonella enterica at different growth stages, and discovered many novel non-coding RNAs and important mRNA regulatory hubs.
Direct RNA-RNA interaction is a main mechanism by which noncoding RNAs exert their functions. In prokaryotic cells, non-coding small RNAs recognize target mRNAs through very short and imperfect basepairing interactions, which are difficult to be predicted and identified using conventional methods. Bacterial non-coding RNAs have important regulatory functions, controlling the expression of many target genes at the post-transcriptional level, and playing crucial regulatory roles in many biological processes, such as bacterial infection, antibiotic resistance, central metabolism, stress response, quorum sensing, and biofilm formation, etc. The analysis of the RNA-RNA interaction network is of great significance to the study of microbial physiology and the functions of non-coding RNAs.
However, the existing RNA-RNA interactome profiling techniques are complicated, which require fixed cells and numerous in vitro steps such as RNA digestion, repair and ligation, yielding insufficient RNA interactions with low resolution and accuracy.
In this study, the researchers developed a novel in vivo RNA proximity-ligation approach, iRIL-seq (intracellular RNA interaction by ligation and sequencing). In this technology, the researchers induced the expression of T4 RNA ligase in living microbial cells, which leads to the proximity-ligation between interacting RNA molecules in vivo; next enriched the non-coding small RNAs and their interacting transcripts by immunoprecipitation of the major small RNA chaperone Hfq; and finally determined the identity and sequence of the interacting RNA molecules using high-throughput RNA sequencing. The main experimental process can be completed within a single day, thanks to the rapid ligation in vivo and streamlined workflow.
This new technology not only illustrates the atlas of microbial RNA-RNA interaction network, but also identifies RNA-RNA basepairing interactions at the single-nucleotide resolution. Therefore, the iRIL-seq technology provides a simple, fast, accurate and universal technology for analyzing the RNA-RNA interactions in microorganisms, and also proposes a new way to study RNA-RNA interactions in eukaryotic cells.
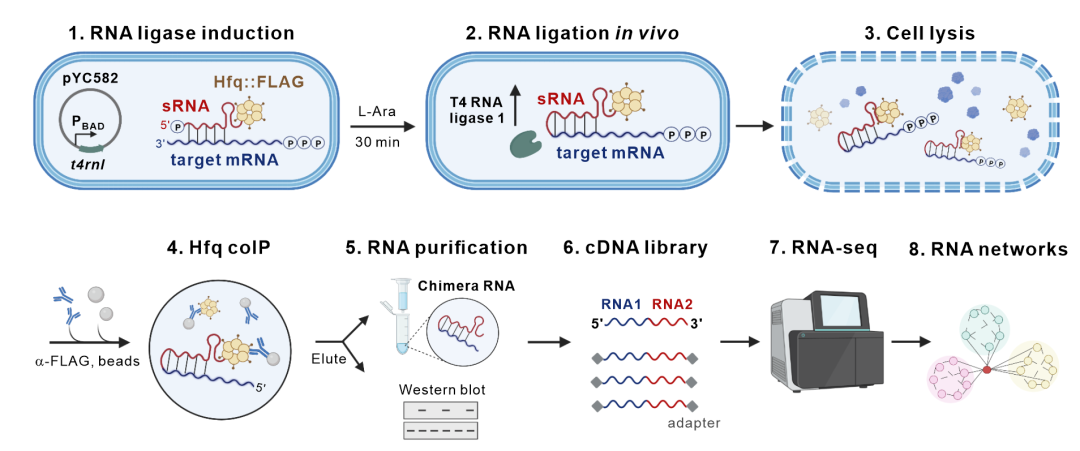
Figure 1. Experimental workflow of iRIL-seq, a novel RNA-RNA interactome profiling technology for live microbial cells.(Image by SIII)
The researchers further mapped for the first time the dynamic RNA-RNA interactomes of bacterial pathogen Salmonella enterica at multiple growth stages, identified more than 2,000 RNA-RNA interactions, and discovered many key mRNA regulatory hubs. The largest mRNA regulatory hub in bacteria to date was shown to be the mRNA encoding the outer membrane porin OmpD, which is regulated by at least 12 different non-coding small RNAs.
Experimental characterization of these small RNAs led to the identification of a novel small RNA FadZ that is cleaved from an mRNA 3'UTR by ribonuclease. The expression of FadZ and its parental fadBA mRNA are regulated by the upstream transcription factors FadR, CRP, and long-chain fatty acid signals. Under long-chain fatty acid conditions, both FadZ sRNA and its target gene ompD are activated by the transcription factor CRP, together constituting a type I incoherent feed-forward loop (I1-FFL). The discovery reveals the post-transcriptional regulatory mechanism of bacteria in response to long-chain fatty acid metabolism in hosts.
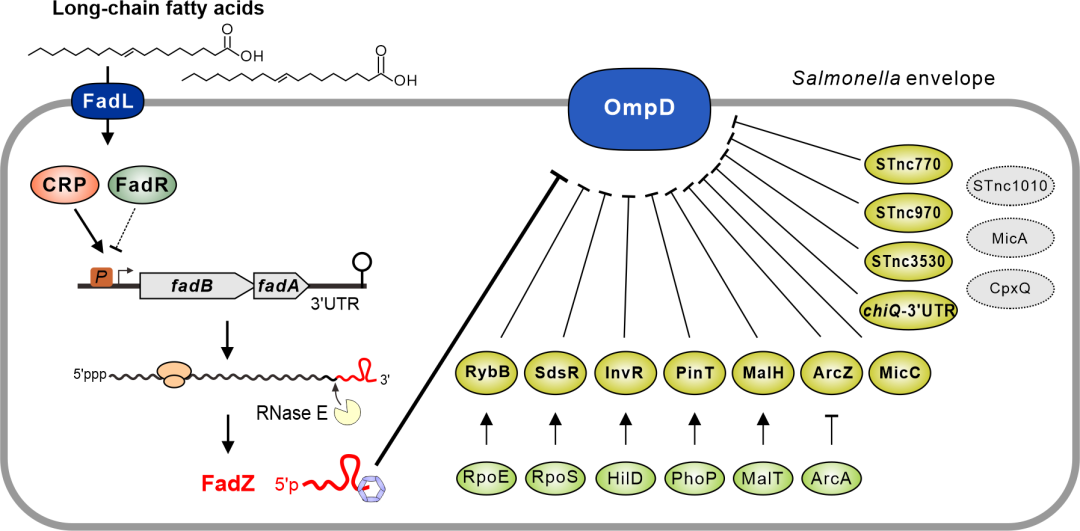
Figure 2. iRIL-seq identifies the mRNA encoding outer-membrane major porin OmpD as the largest mRNA hub in Salmonella. The ompD mRNA is regulated by 12 different small noncoding RNAs, including a novel 3’UTR-derived sRNA FadZ.(Image by SIII)
Contact
CHAO Yanjie
Shanghai Institute of Immunity and Infection, CAS
E-mail: yjchao@siii.cas.cn
Homepage: http://yanjiechao.wordpress.com
Reference: https://doi.org/10.1038/s41467-023-43632-1
Link of CAS: https://english.cas.cn/newsroom/research_news/life/202312/t20231208_653812.shtml

