Kaposi’s sarcoma-associated herpesvirus (KSHV), known as human herpesvirus 8 (HHV8), is the etiological agent of Kaposi’s sarcoma (KS), primary effusion lymphoma (PEL) and multicentric Castelman’s disease (MCD). KSHV has developed a unique mechanism to subvert host antiviral immune responses by encoding four homologues of cellular interferon regulatory factor (vIRF1~4, vIRFs) with a cluster of KSHV open reading frames K9, K11/11.1, K10.5/10.6, K10 respectively. vIRFs have been demonstrated as effective inhibitors of interferon signaling and regulators of cellular oncogenic pathways, which may contribute to KSHV infection as well as pathogenesis.
From the previous research, vIRF2 appears to be able to inhibit both early and later steps of the antiviral IFN signaling pathway. The vIRF2 can function as modulator of the innate immune response through negatively regulating the transactivation of the ISRE promoter by IRF-1 as well as activation of IFN reporter promoter. Moreover, vIRF2 targets components of the interferon-stimulated gene factor 3 (ISGF3) complexes, STAT1 and IRF-9, which results in the inhibition of ISG expression. Previous studies have shown that vIRF2 were unable to bind to the consensus DNA sequence of the human IRFs. The inhibitory role of vIRF2 in regulating IFN signaling seemed to rely on its interaction with other regulatory factors, but not direct transcriptional regulation.
The research was supervised by Prof. Ke Lan and was completed by Haidai Hu and Jiazheng Dong cooperated with Prof. Yuhui Dong in IHEP(Institute of High Energy Physics, Chinese Academy of Sciences).Researchers performed the genome-wide vIRF2-binding sites mapping in human genome using chromatin immunoprecipitation coupled to high-throughput sequencing (ChIP-seq). vIRF2 was capable of binding to the promoter regions of 100 putative target genes. Importantly, researchers confirmed vIRF2 can specifically interact with the promoters of the genes encoding PIK3C3, HMGCR and HMGCL are associated with autophagosomes formation or tumor progression and metastasis, regulate their transcriptions in vivo. The crystal structure of vIRF2 DNA-binding domain (DBD) showed variable loop conformations and different positive charge distribution from vIRF1 to cellular IRFs that are associated with DNA-binding specificities. Structure-based mutagenesis revealed Arg82 and Arg85 are required in vitro DNA binding activity of vIRF2DBD, and can abolish the transcription regulation function of vIRF2 on the promoter reporter activity of PIK3C3, HMGCR and HMGCL. The study provided unique insights into the DNA binding potency of vIRF2 and suggested that vIRF2 could act as a transcription factor of its target genes in host antiviral immune response.
The article entitled “Genome-wide mapping of the binding sites and structural analysis of KSHV vIRF2 reveal it is a DNA-binding transcription factor” was published online in Journal of Virology on November 4.
The study was supported by the key project of the Natural Science Foundation of China and the National Basic Research Program (973).
The link for this article: http://www.ncbi.nlm.nih.gov/pubmed/26537687
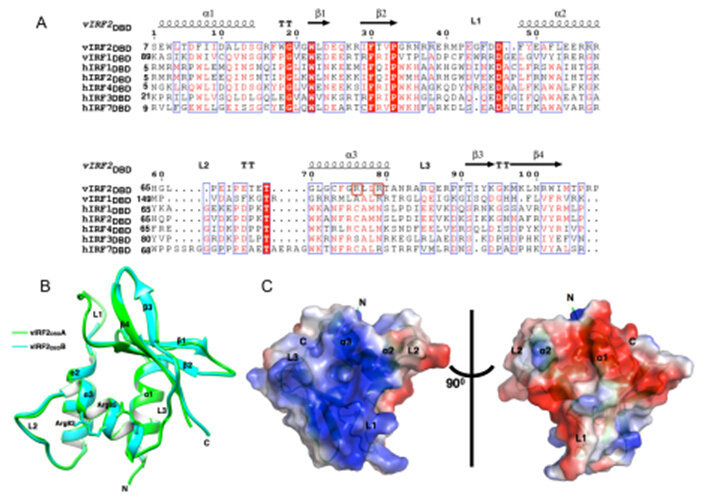
Fig. 3. Structural characteristics of vIRF2DBD. (A) Structure-based sequence alignment for vIRF2DBD with those in vIRF1 and human IRFs DBD. (B) Overall view of vIRF2DBD structure shown as cartoon. (C) The molecular surface representation of vIRF2DBD colored by their local electrostatic potential

