On April 17, the international academic journal Journal of Virology published an article online entitled “Kaposi's Sarcoma-Associated Herpesvirus-Encoded LANA Interacts with Host KAP1 to Facilitate Establishment of Viral Latency”by Prof. Ke Lan’s lab at Institut Pasteur of Shanghai, Chinese Academy of Sciences.
Kaposi’s sarcoma-associated herpesvirus (KSHV) typically displays two different phases in life cycle, including the default latent phase as well as the lytic phase. There is a short period of lytic gene expression at the early stage of KSHV primary infection. The factors involved in the shutdown process of lytic gene expression are poorly identified.
From the previous research, it showed that the latency-associated nuclear antigen (LANA) encoded by KSHV played an important role in the establishment of viral latency. LANA protein is essential for the replication and persistence of viral episome during latent infection. Deleting LANA from the KSHV genome results in the loss of the viral episome, and enhances lytic gene expression. LANA can inhibit RTA expression by repressing its promoter. However, how LANA represses the transcriptional activity of RTA promoter remains largely unknown.
The researchers identified a host protein, Krüppel-associated box domain associated protein-1 (KAP1) that bound to LANA through TAP-MS (tandem affinity purification-mass spectrometry) method, KAP1 was shown to change epigenetic state through recruiting histone deacetylase and methyltransferase complex. The researchers validated the interaction between LANA and KAP1 as well as their colocalization in the nucleus, and also mapped out LANA interacted with both the N- and C-terminal domains of KAP1. Based on the determined interface of LANA-KAP1 interaction, it was proved that LANA recruited KAP1 to the RTA promoter region of the KSHV genome and KAP1 was involved in transcriptional repression by LANA. To determine whether LANA recruited KAP1 to other regions of the KSHV genome, the researchers mapped the whole genome-wide binding sites of LANA and KAP1 by ChIP-seq, and found multiple co-occupation sites of LANA and KAP1 on the whole KSHV genome. Moreover, LANA-recruited KAP1 was demonstrated to play a critical role in the shutdown of lytic gene expression during the early stage of KSHV primary infection. This study revealed the mechanism of transcriptional repression by LANA during KSHV primary infection, providing new insights into the process of KSHV latency establishment.
The research was supervised by Prof. Ke Lan and was completed by Ph.D student Rui Sun and other group members. The study received technical support from the Omics Core, CAS-MPG Partner Institute for Computational Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. The study was supported by the key project of the Natural Science Foundation of China, the National Basic Research Program (973) and other projects.
The link for this article: http://www.ncbi.nlm.nih.gov/pubmed/24741090
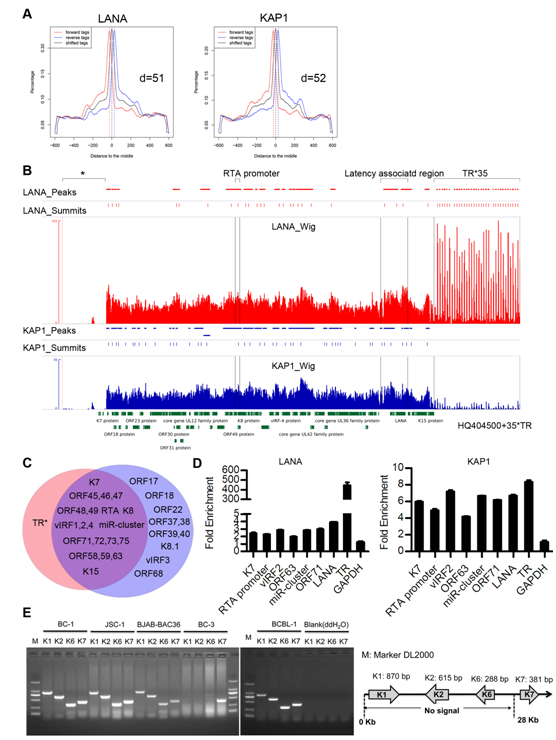
Fig.6. LANA and KAP1 had multiple co-occupation sites on the KSHV genome. (A) Peak models of LANA and KAP1 built by MACS based on the human genome alignment. (B) Illustration of LANA and KAP1 binding sites on the whole KSHV genome. (C) Illustration of regions occupied by LANA(red)and KAP1(blue).(P<10–5) (D) Validation of ChIP-seq results by qPCR. (E) Validation of possible deletion or big sequence variation in the KSHV genome of BC-3 cells by PCR.

